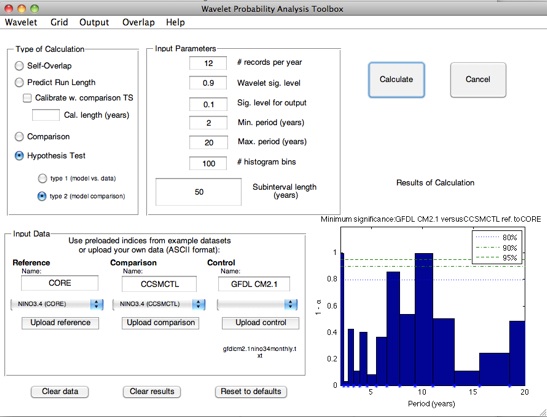
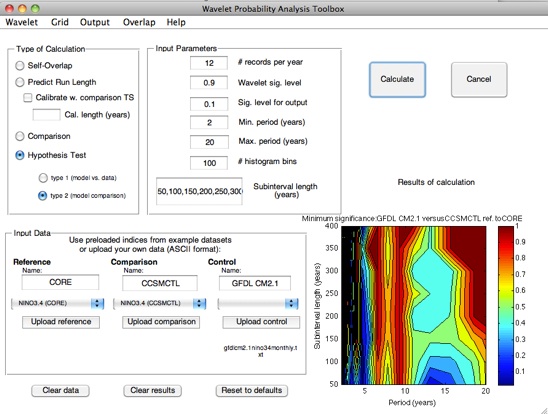
For some applications, the quantity of interest is the degree of self-consistency shown by a model run. This is the case for long control integrations, for example; here it is essential to ensure that you are capturing as much of the full range of variability in the system as possible.
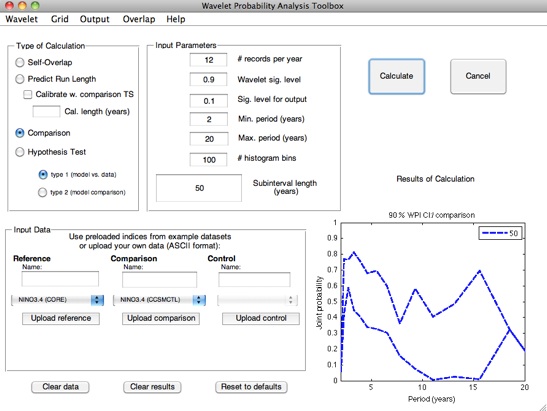
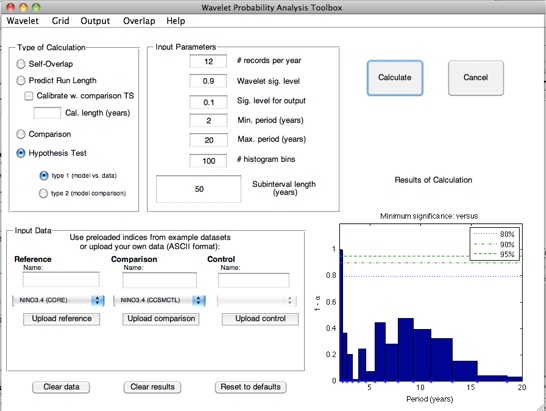
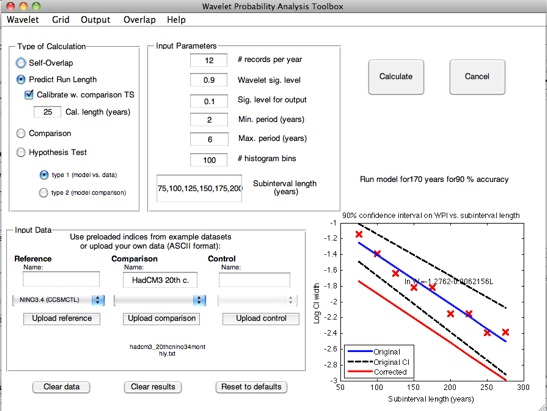
In terms of wavelet probability analysis, answering this question requires a self-overlap calculation. Stevenson et al. (2010) showed that the width of the confidence interval on self-overlap WPI has an exponential dependence on the length of the subinterval of the time series used; additionally, this dependence has a statistically identical slope between different climate models. This behavior is the basis for the prediction algorithm used by the wavelet probability analysis scripts.
Using the GUI:
-
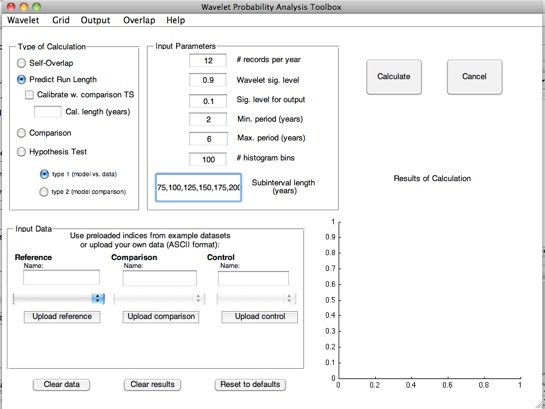
1.Set the “Calculation Type” to “Predict Run Length”. This will change the default subinterval length from 50 years to a specified array of values ranging from 75 to 275 years, which will become the input for the regression. Subinterval lengths may be changed at any time before pressing the “Calculate” button.
-
2.Specify the dataset for regression: this will be entered as the reference time series.
One may imagine that using a relatively short (1-200 year) control integration of a model will not give a very accurate
regression slope; yet this may be the only control run available for that particular model. In that case, using one of the pre-
loaded, longer datasets may be more useful. You specify this by using the drop-down menu to select the dataset you would like
to use.
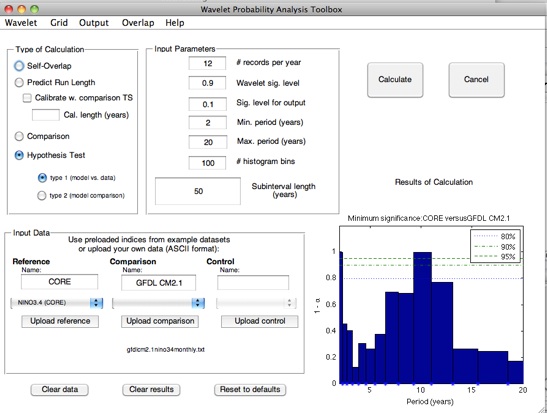
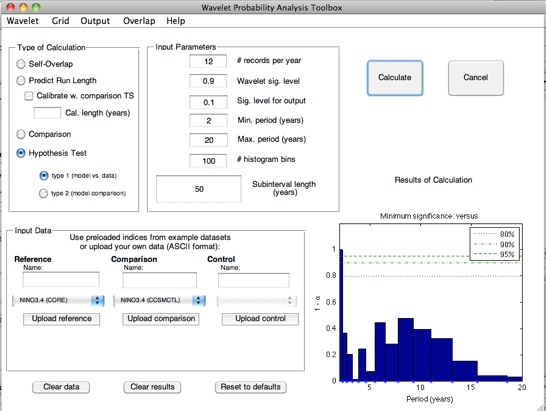
Stevenson et al. (2010) points out that although the slope of the WPI confidence interval regression seems to hold across
climate models, the intercept is shifted up or down depending on the total length of the dataset being used to generate the
confidence intervals. The GUI allows you to automatically calibrate for this effect, using a time series of arbitrary length: select
the “Calibrate w. comparison TS” checkbox to do so. You will then need to input your desired time series as the “comparison”.
What actually happens when you calibrate using the comparison dataset is that the regression slope is calculated using the
specified pre-loaded dataset. The program then takes the input subinterval length from the “Cal length” text box, and does a
self-overlap calculation at that subinterval length on the shorter, input reference. The width of the latter confidence interval is
then used to apply a constant offset to the remaining data.